library(tidyverse)
library(cluster)
library (janitor)
library(factoextra)
library (patchwork)
library (readxl)
library (fpc)
library (gt)
library (PerformanceAnalytics)Não Supervisionado - Cluster
Base de Dados
# Selecionar variáveis quantitativas
df <- mtcars |>
select (mpg, disp, hp, drat, wt, qsec) |>
# Fazer o z-score (padronizacao)
scale() |> data.frame()Correlações
chart.Correlation(df, histogram = TRUE, method = "pearson")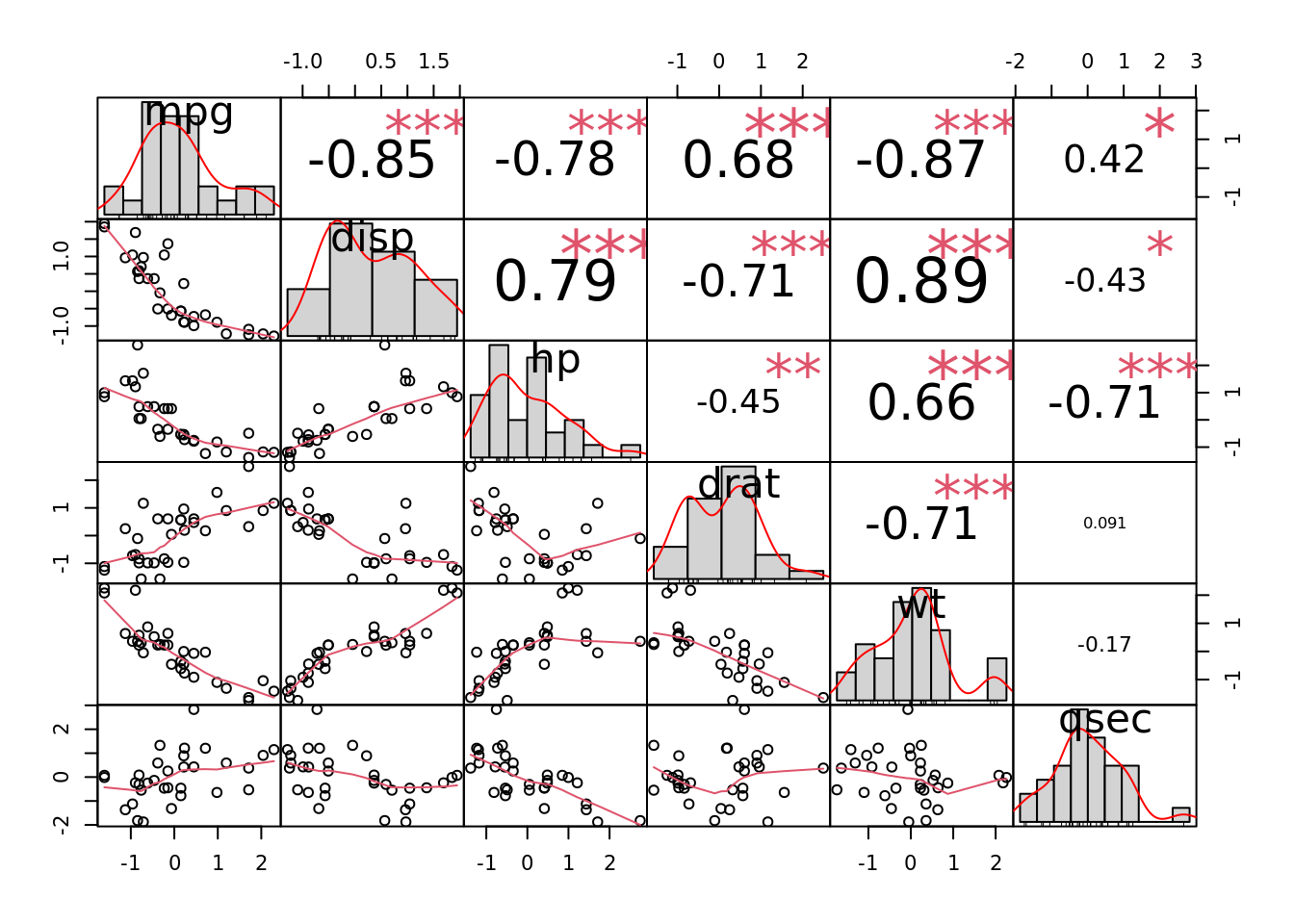
DataExplorer::plot_correlation(df)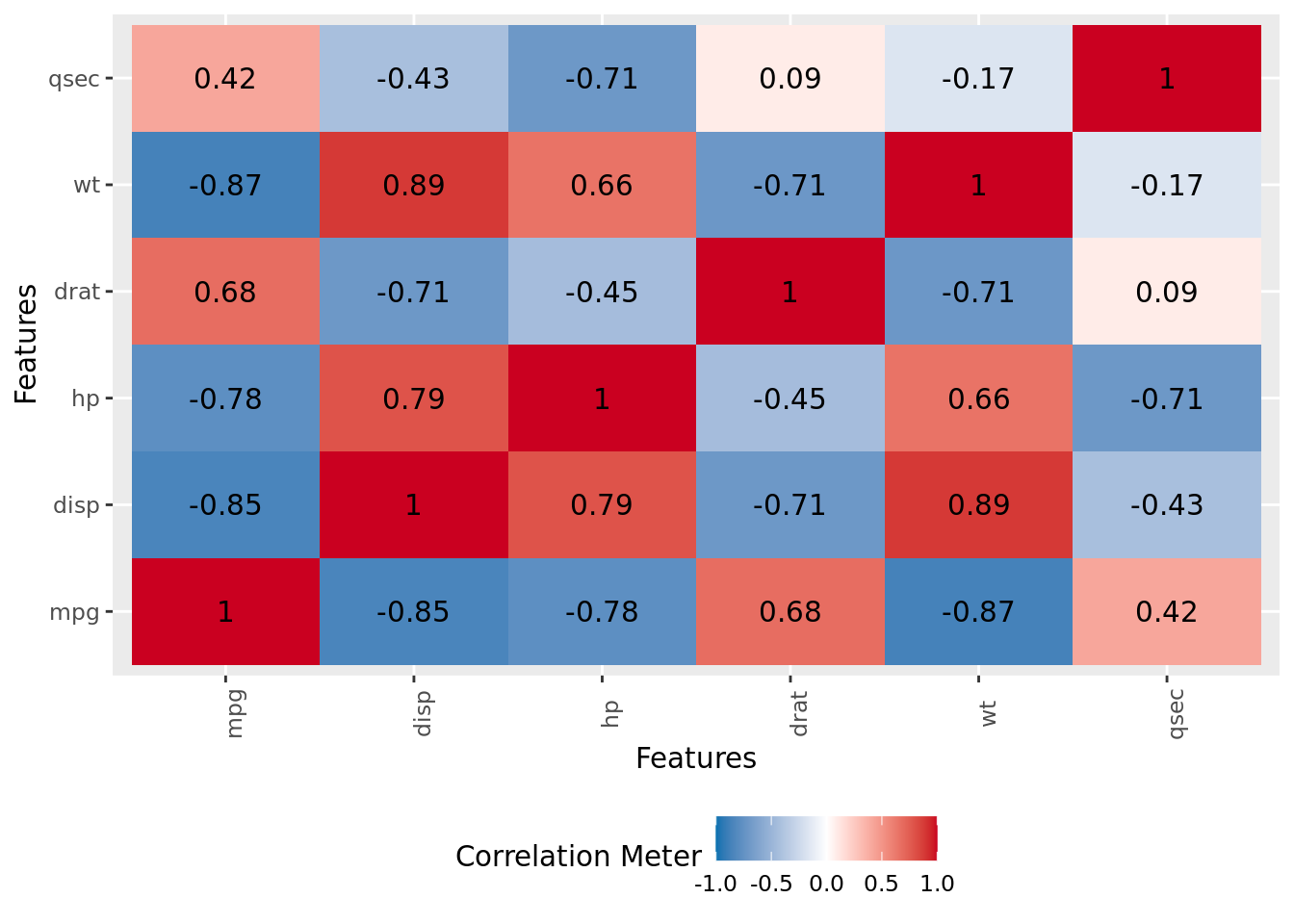
Método Hierárquico
Usando hclust
#calcular as distancias da matriz utilizando a distancia euclidiana
distancia <- dist(df, method = "euclidean")
#Calcular o Cluster
clusterh <- hclust(distancia, method = "single" )
# Dendrograma
plot(clusterh, cex = 0.6, hang = -1)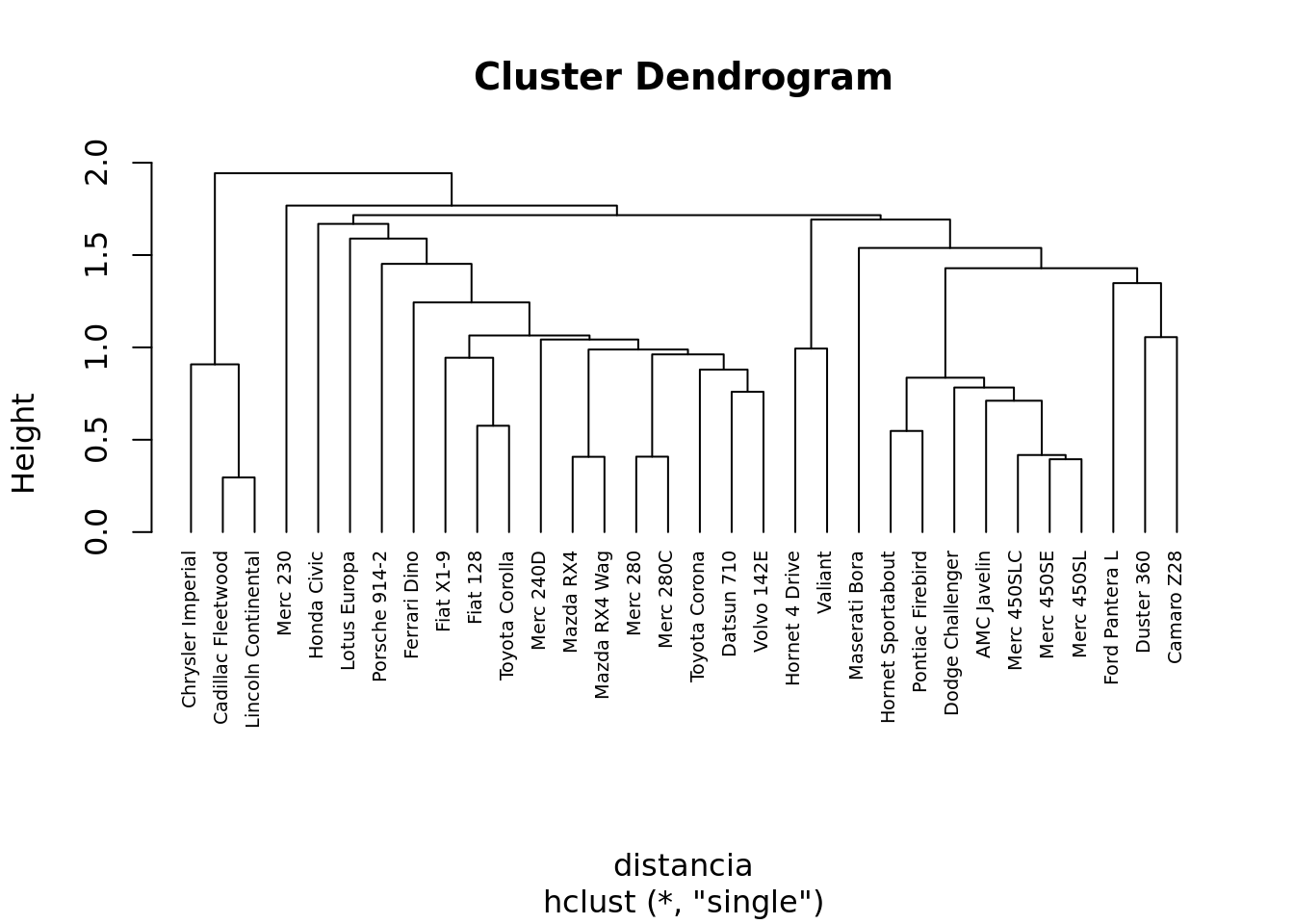
#Metodo Elbow
fviz_nbclust(df, FUN = hcut, method = "wss")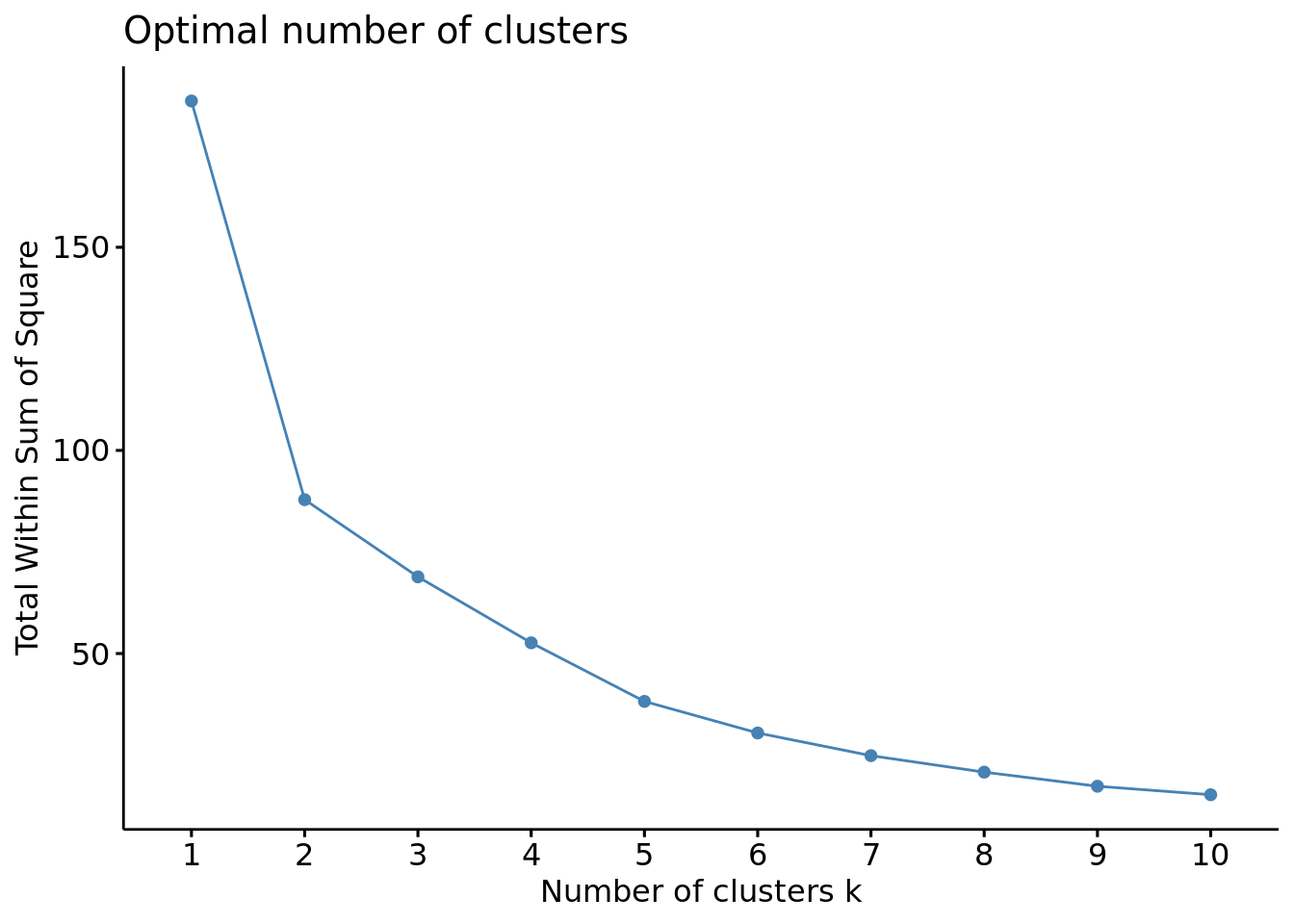
#criando 5 grupos
clusterh_grupos <- cutree(clusterh, k = 5)
table(clusterh_grupos)clusterh_grupos
1 2 3 4 5
15 2 11 1 3 #transformando em data frame a saida do cluster
grupo_hierarquico <- data.frame(clusterh_grupos) |>
dplyr::rename (cluster = last_col())
df_final <- bind_cols(mtcars, grupo_hierarquico)
df_final_EDA <- df_final |> pivot_longer(cols = c(1,3:7))
plot_final_EDA <- df_final_EDA |>
ggplot(aes(x=cluster, y=value, color= as_factor(cluster)))+
#geom_jitter(alpha = 0.5, size = 6, width = 0.1)+
geom_boxplot()+
facet_grid(scales = "free_x", cols = vars(name))
plot_final_EDA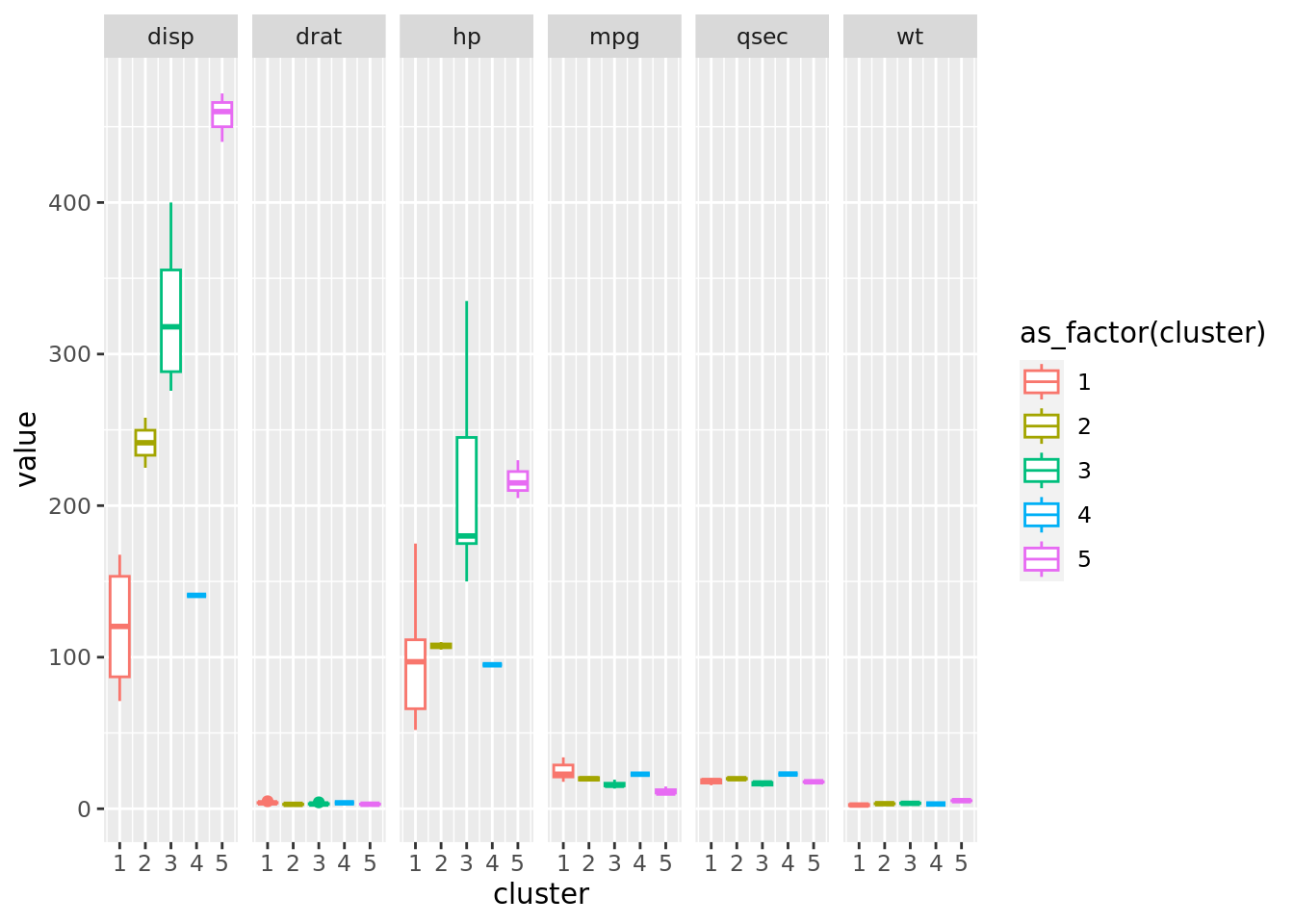
Usando agnes
#Comparar os tipos de link
m <- c( "average", "single", "complete", "ward")
names(m) <- c( "average", "single", "complete", "ward")
map(m, ~agnes(df, method = .)$ac)$average
[1] 0.7659485
$single
[1] 0.5631059
$complete
[1] 0.867514
$ward
[1] 0.9320082#Calcular o Cluster
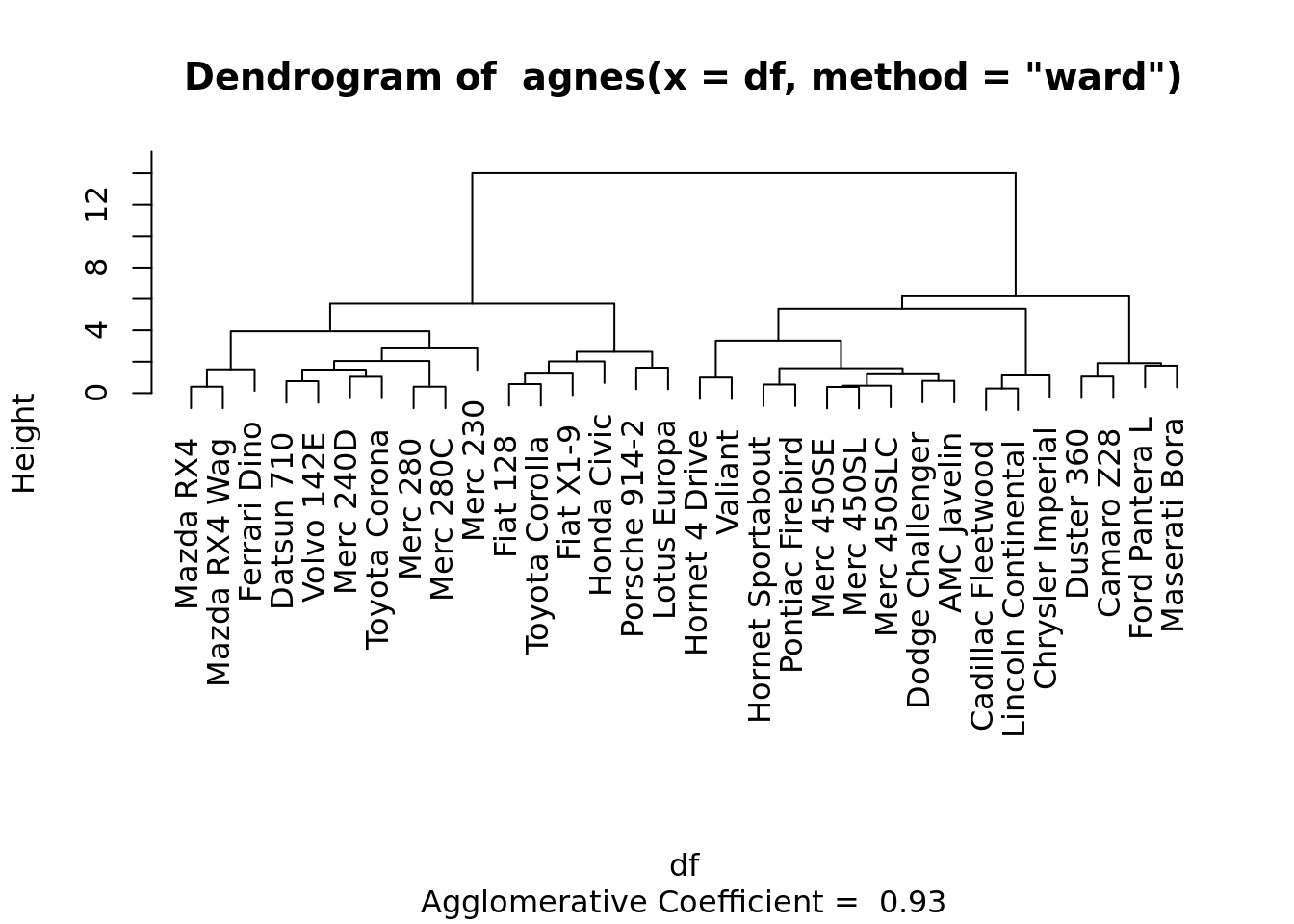
clustera <- agnes(df, method = "ward" )
# Dendrograma
pltree(clustera, cex = 0.6, hang = -1)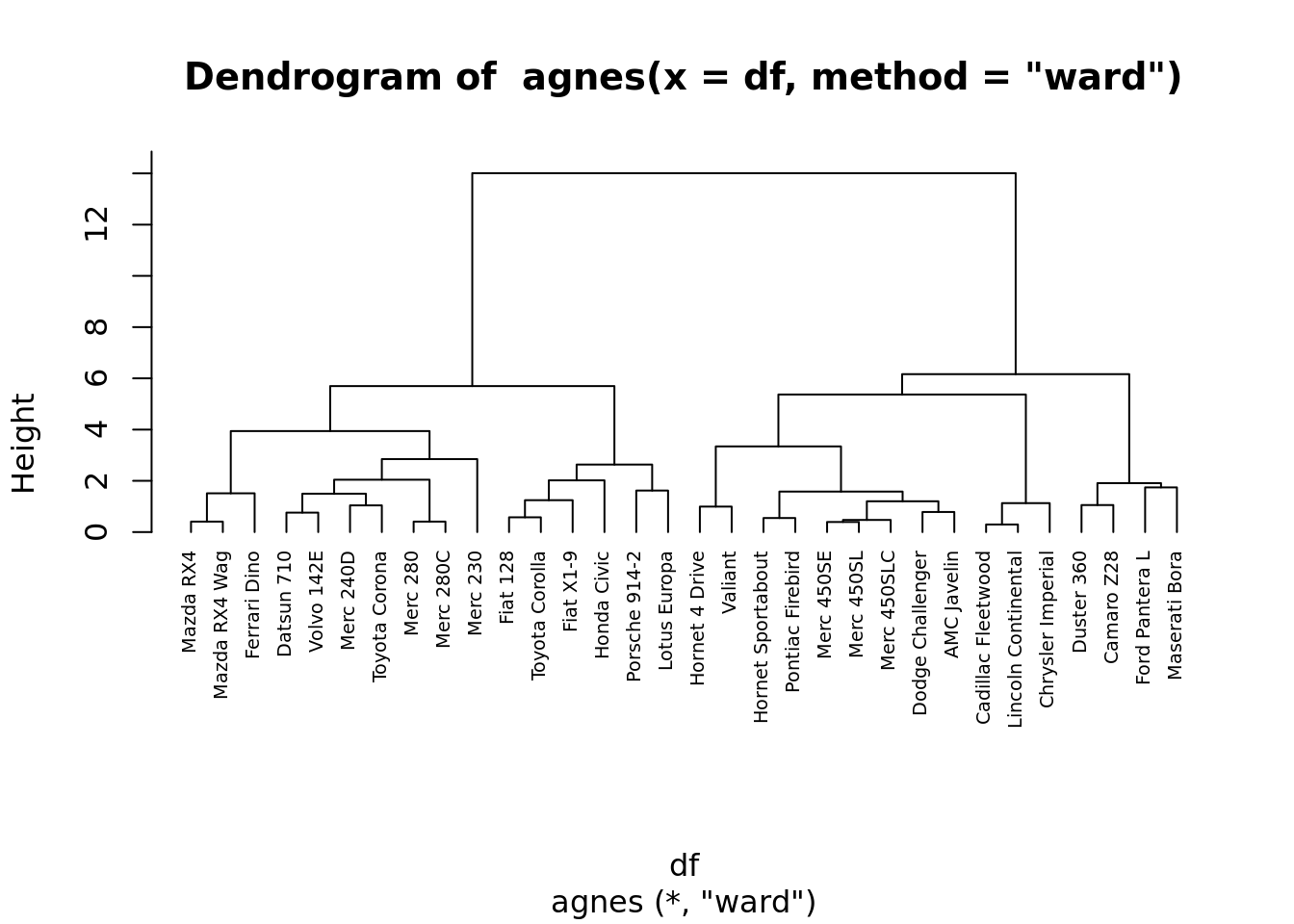
#Agnes plot ( dendograma e banner plot)
plot(clustera)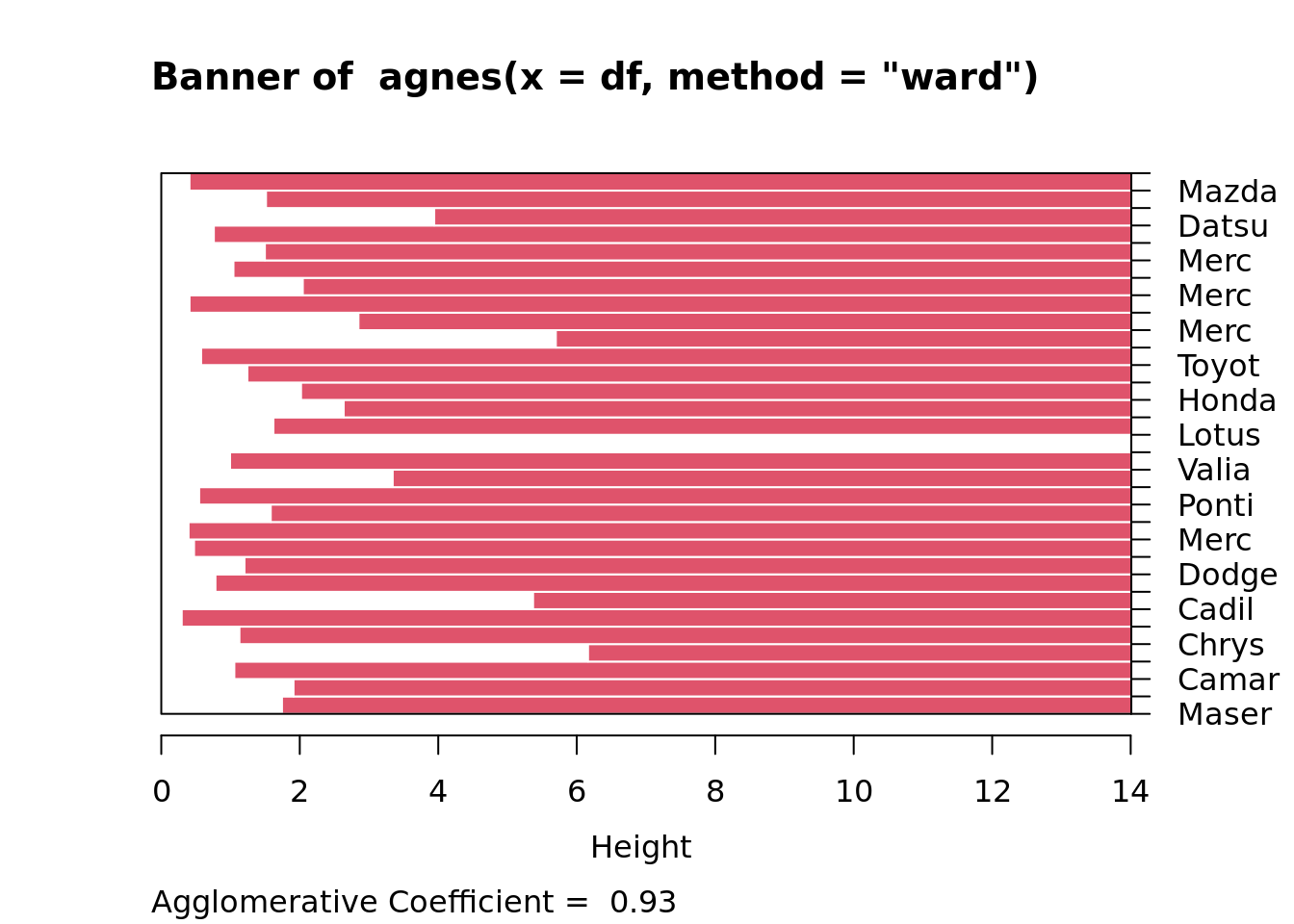
#criando 5 grupos
clustera_grupos <- cutree(clustera, k = 5)
table(clustera_grupos)clustera_grupos
1 2 3 4 5
10 9 4 3 6 #transformando em data frame a saida do cluster
grupo_hierarquico_a <- data.frame(clustera_grupos) |>
dplyr::rename (cluster = last_col())
df_final <- bind_cols(mtcars, grupo_hierarquico)
df_final_EDA <- df_final |> pivot_longer(cols = c(1,3:7))
plot_final_EDA <- df_final_EDA |>
ggplot(aes(x=cluster, y=value, color= as_factor(cluster)))+
#geom_jitter(alpha = 0.5, size = 6, width = 0.1)+
geom_boxplot()+
facet_grid(scales = "free_x", cols = vars(name))
plot_final_EDA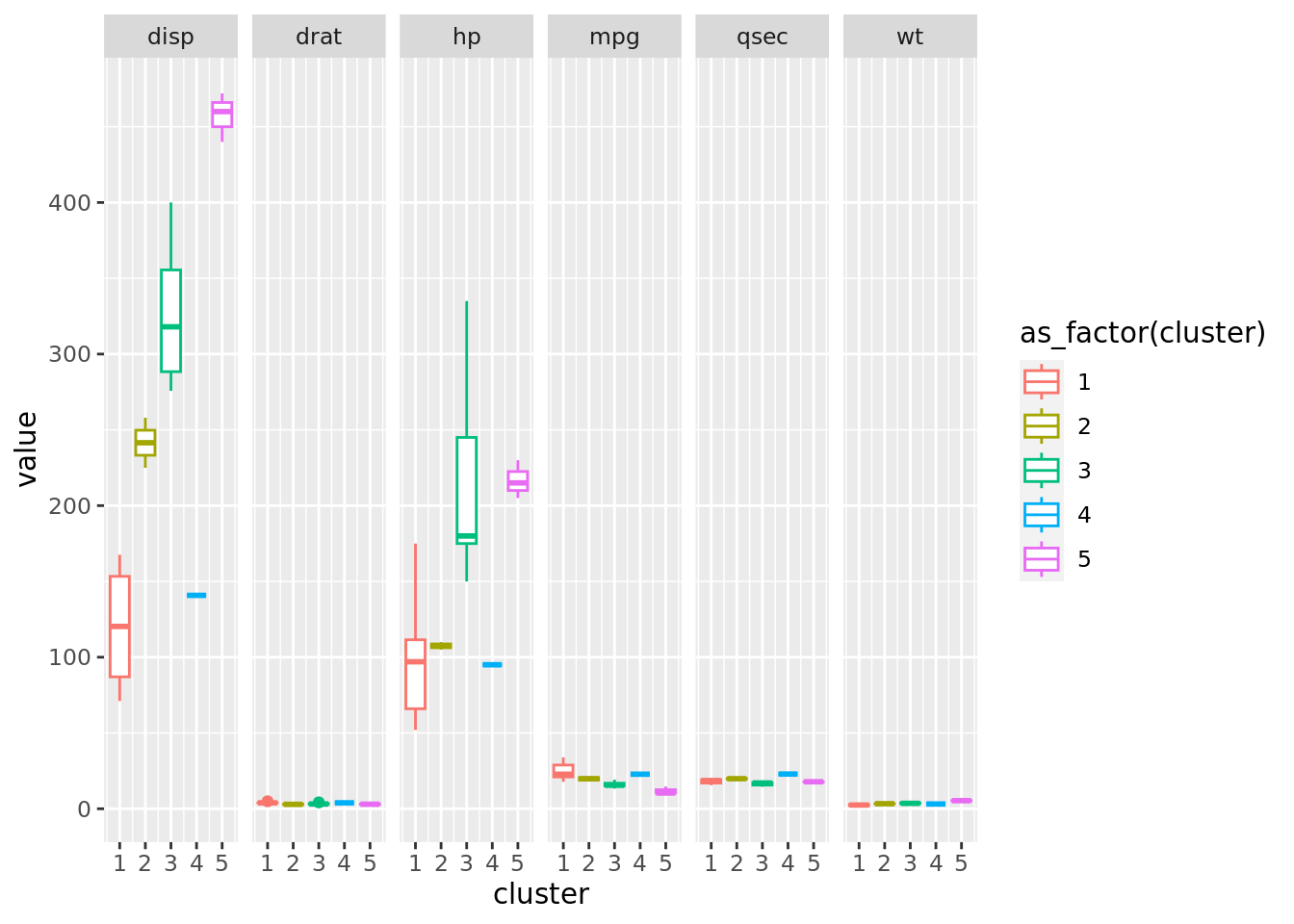
Warning
Veja que agnes(*, method=“ward”) corresponde a hclust(*, “ward.D2”).
Método Não-Hierárquico
K-Means
O número de cluster (k) precisa ser dado
#Método Elbow
fviz_nbclust(df, FUN = hcut, method = "wss")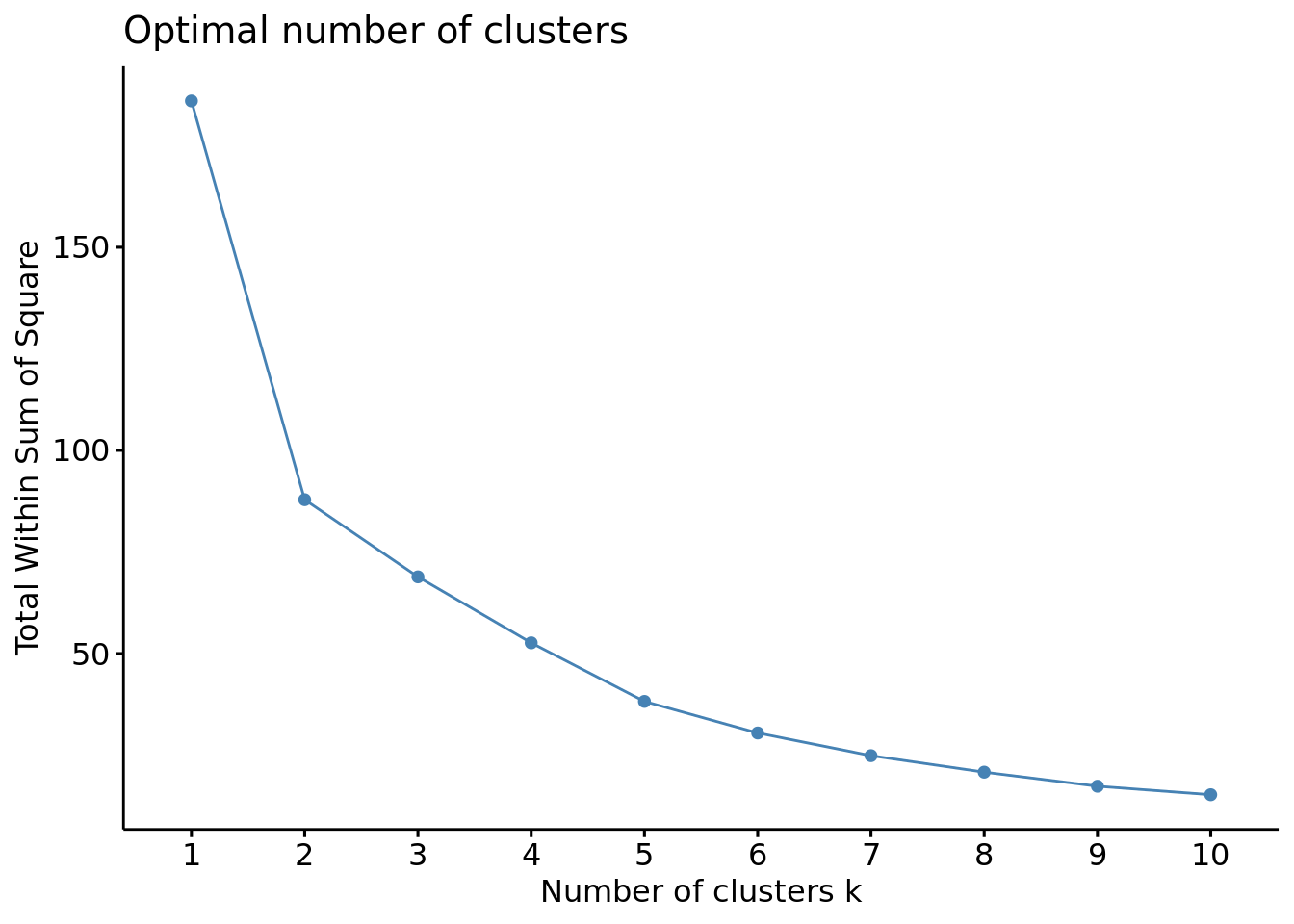
#Gerando clsuters com 3, 4, 5 e 6 grupos
k3 <- kmeans(df, centers = 3)
k4 <- kmeans(df, centers = 4)
k5 <- kmeans(df, centers = 5)
k6 <- kmeans(df, centers = 6)
lista <- list(k3,k4,k5,k6)
names(lista) <- c("k3", "k4", "k5", "k6")
G <- map(lista, ~fviz_cluster(., geom = "point", data = df) +
ggtitle(paste(length(
(pluck(., 7))),"clusters")))
G$k3 + G$k4 + G$k5 + G$k6 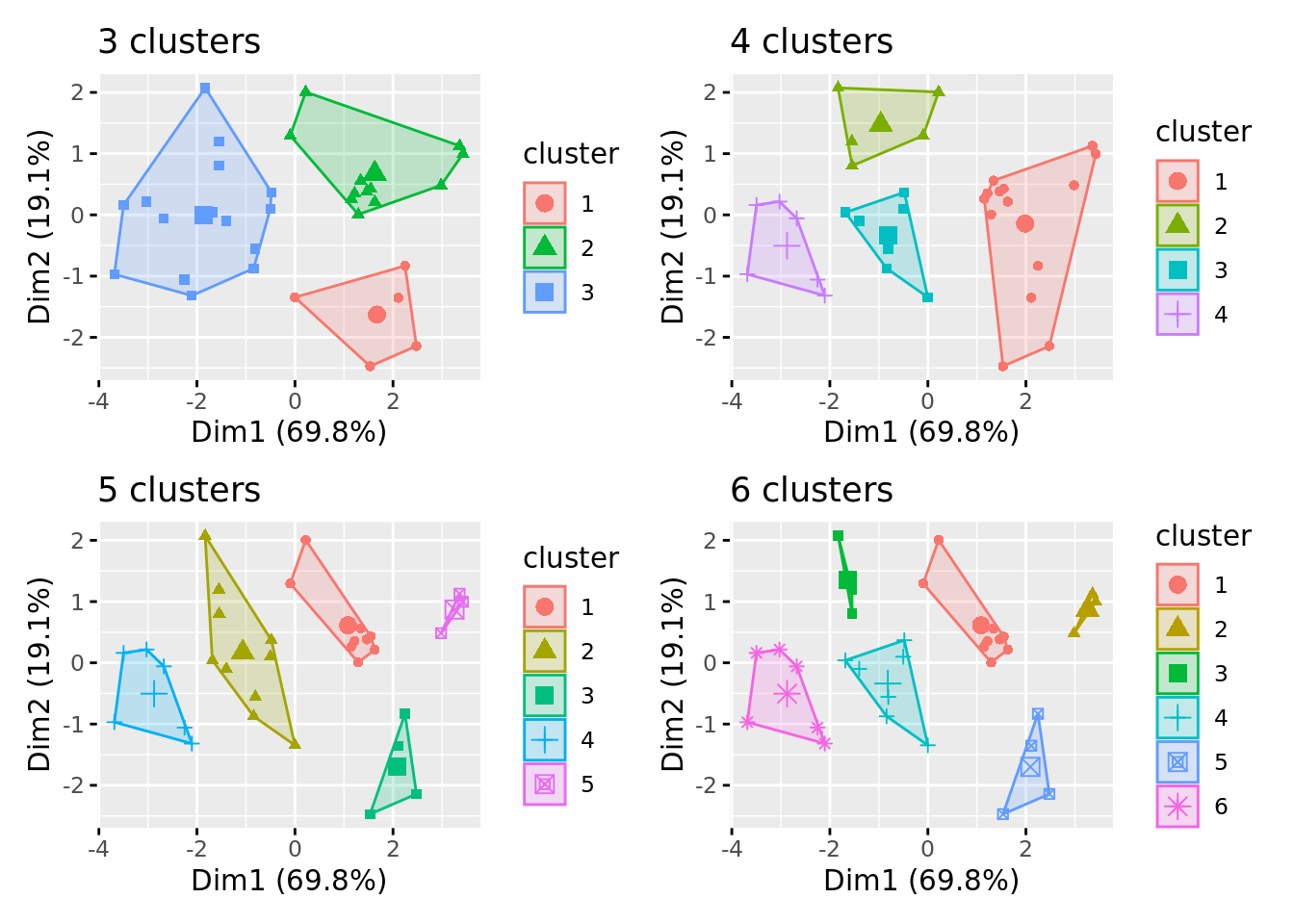
Escolhendo 5 grupos
grupo_kmeans <- data.frame(k5$cluster) |>
dplyr::rename (cluster = last_col())
df_final_kmeans <- bind_cols(mtcars, grupo_kmeans)
df_final_kmeans_EDA <- df_final_kmeans |> pivot_longer(cols = c(1,3:7))
plot_final_kmeans_EDA <- df_final_kmeans_EDA |>
ggplot(aes(x=cluster, y=value, color= as_factor(cluster)))+
#geom_jitter(alpha = 0.5, size = 6, width = 0.1)+
geom_boxplot()+
facet_grid(scales = "free_x", cols = vars(name))
plot_final_kmeans_EDA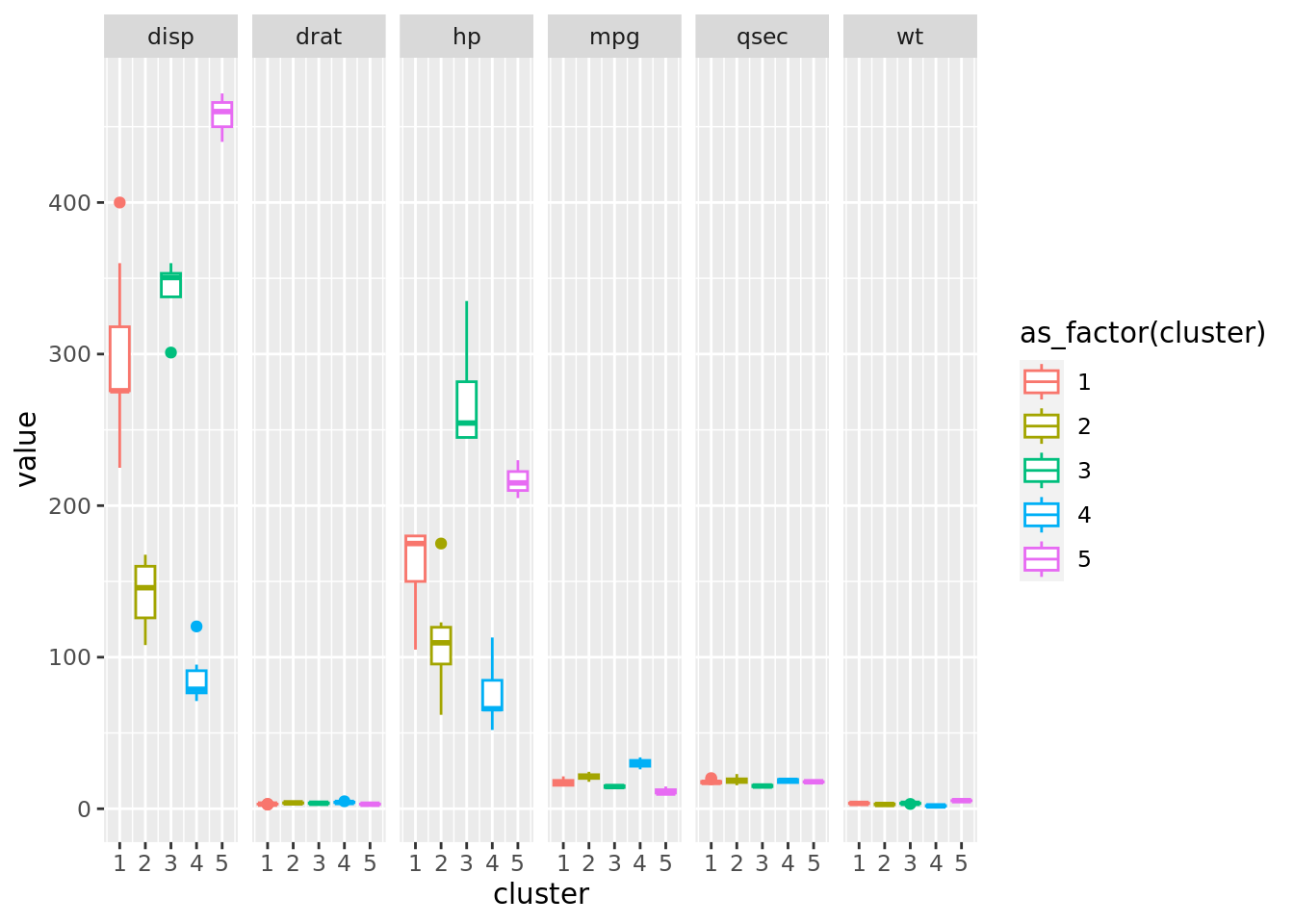
Visualizando os dados com os respetivos clsuters k-means
df_final_kmeans |>
group_by(cluster) |>
gt(rowname_col = "name") |>
tab_stubhead(label ="Modelo") |>
tab_options(row.striping.include_table_body = TRUE) |>
text_transform(locations = cells_row_groups(),
fn = function(x) {
paste0("Cluster: ", x)
})| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb |
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster: 2 | ||||||||||
| 21.0 | 6 | 160.0 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 21.0 | 6 | 160.0 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 22.8 | 4 | 108.0 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| 24.4 | 4 | 146.7 | 62 | 3.69 | 3.190 | 20.00 | 1 | 0 | 4 | 2 |
| 22.8 | 4 | 140.8 | 95 | 3.92 | 3.150 | 22.90 | 1 | 0 | 4 | 2 |
| 19.2 | 6 | 167.6 | 123 | 3.92 | 3.440 | 18.30 | 1 | 0 | 4 | 4 |
| 17.8 | 6 | 167.6 | 123 | 3.92 | 3.440 | 18.90 | 1 | 0 | 4 | 4 |
| 21.5 | 4 | 120.1 | 97 | 3.70 | 2.465 | 20.01 | 1 | 0 | 3 | 1 |
| 19.7 | 6 | 145.0 | 175 | 3.62 | 2.770 | 15.50 | 0 | 1 | 5 | 6 |
| 21.4 | 4 | 121.0 | 109 | 4.11 | 2.780 | 18.60 | 1 | 1 | 4 | 2 |
| Cluster: 1 | ||||||||||
| 21.4 | 6 | 258.0 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| 18.7 | 8 | 360.0 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| 18.1 | 6 | 225.0 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
| 16.4 | 8 | 275.8 | 180 | 3.07 | 4.070 | 17.40 | 0 | 0 | 3 | 3 |
| 17.3 | 8 | 275.8 | 180 | 3.07 | 3.730 | 17.60 | 0 | 0 | 3 | 3 |
| 15.2 | 8 | 275.8 | 180 | 3.07 | 3.780 | 18.00 | 0 | 0 | 3 | 3 |
| 15.5 | 8 | 318.0 | 150 | 2.76 | 3.520 | 16.87 | 0 | 0 | 3 | 2 |
| 15.2 | 8 | 304.0 | 150 | 3.15 | 3.435 | 17.30 | 0 | 0 | 3 | 2 |
| 19.2 | 8 | 400.0 | 175 | 3.08 | 3.845 | 17.05 | 0 | 0 | 3 | 2 |
| Cluster: 3 | ||||||||||
| 14.3 | 8 | 360.0 | 245 | 3.21 | 3.570 | 15.84 | 0 | 0 | 3 | 4 |
| 13.3 | 8 | 350.0 | 245 | 3.73 | 3.840 | 15.41 | 0 | 0 | 3 | 4 |
| 15.8 | 8 | 351.0 | 264 | 4.22 | 3.170 | 14.50 | 0 | 1 | 5 | 4 |
| 15.0 | 8 | 301.0 | 335 | 3.54 | 3.570 | 14.60 | 0 | 1 | 5 | 8 |
| Cluster: 5 | ||||||||||
| 10.4 | 8 | 472.0 | 205 | 2.93 | 5.250 | 17.98 | 0 | 0 | 3 | 4 |
| 10.4 | 8 | 460.0 | 215 | 3.00 | 5.424 | 17.82 | 0 | 0 | 3 | 4 |
| 14.7 | 8 | 440.0 | 230 | 3.23 | 5.345 | 17.42 | 0 | 0 | 3 | 4 |
| Cluster: 4 | ||||||||||
| 32.4 | 4 | 78.7 | 66 | 4.08 | 2.200 | 19.47 | 1 | 1 | 4 | 1 |
| 30.4 | 4 | 75.7 | 52 | 4.93 | 1.615 | 18.52 | 1 | 1 | 4 | 2 |
| 33.9 | 4 | 71.1 | 65 | 4.22 | 1.835 | 19.90 | 1 | 1 | 4 | 1 |
| 27.3 | 4 | 79.0 | 66 | 4.08 | 1.935 | 18.90 | 1 | 1 | 4 | 1 |
| 26.0 | 4 | 120.3 | 91 | 4.43 | 2.140 | 16.70 | 0 | 1 | 5 | 2 |
| 30.4 | 4 | 95.1 | 113 | 3.77 | 1.513 | 16.90 | 1 | 1 | 5 | 2 |
DBSCAN
dbscan <- fpc::dbscan(df, eps = .78, MinPts = 3)
grupo_dbscan <- data.frame(dbscan$cluster) |>
dplyr::rename (cluster = last_col())
df_final_dbscan <- bind_cols(mtcars, grupo_dbscan)
df_final_dbscan_EDA <- df_final_dbscan |> pivot_longer(cols = c(1,3:7))
plot_final_dbscan_EDA <- df_final_dbscan_EDA |>
ggplot(aes(x=cluster, y=value, color= as_factor(cluster)))+
#geom_jitter(alpha = 0.5, size = 6, width = 0.1)+
geom_boxplot()+
facet_grid(scales = "free_x", cols = vars(name))
plot_final_dbscan_EDA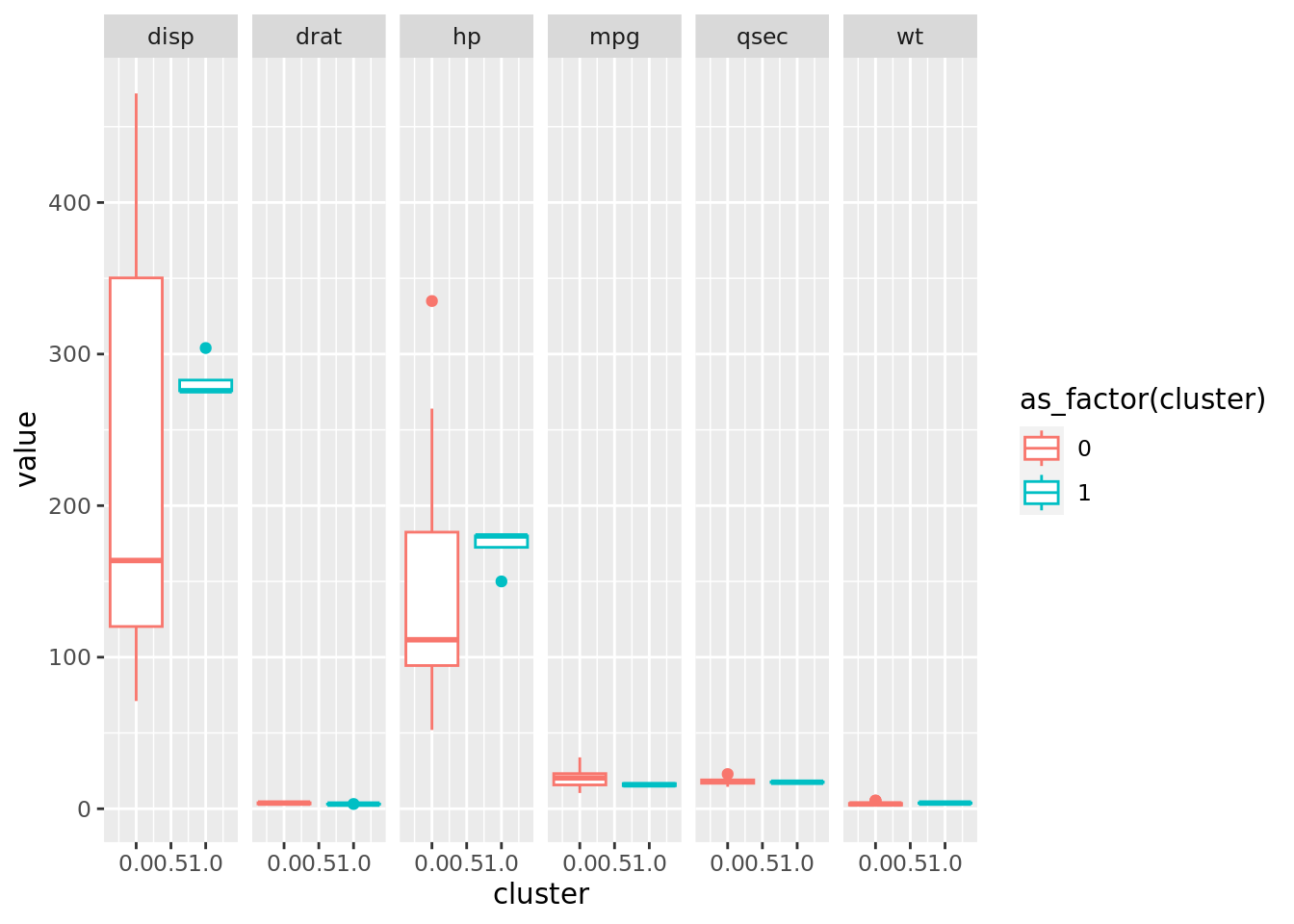
#Outros plots:
plot(dbscan, df)